Mycobacterium phage Salili
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Salili | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Denali Smith, Sarah Nichols, Julian Niewiaroski |
| Year Found | 2016 |
| Location Found | Orono, ME USA |
| Finding Institution | University of Maine, Honors College |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | Not entered |
| GPS Coordinates | 44.905 N, 68.6668 E Map |
| Discovery Notes | Came from dirt on the Umaine bike path, directly at the fork in the paths, behind the tennis courts. The dirt was very compacted, very wet and very full of plants (like roots, leaves and stems). It was a warm day when this was collected. |
| Naming Notes | This sample was worked on by Sarah Nichols, Julian Niewiaroski and Denali Smith. The name Salili is a combination of all three of our first names. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | L |
| Subcluster | -- |
| Cluster Life Cycle | Temperate |
| Lysogeny Notes | The Lysogeny of Salili is 76.1% |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
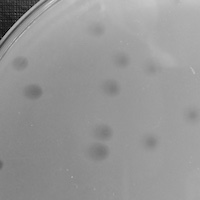
| Plaque Notes | Creates turbid plaques. During assays the plaques have a huge range in sizes. During streaking/isolation, the plaques are typically on the small side. Occasionally plaques with clear centers but turbid outsides were observed. |
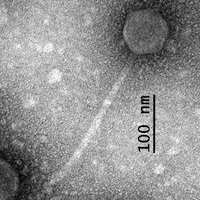
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Pitt Freezer Box# | 44 |
| Pitt Freezer Box Grid# | F10 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |