
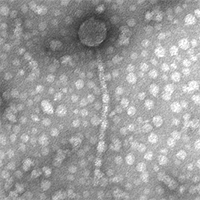
| Detailed Information for Phage Scap1 |
| Discovery Information |
| Isolation Host | Streptomyces scabiei RL-34 |
| Found By | Steven Caruso |
| Year Found | 2016 |
| Location Found | Kottayam, India |
| Finding Institution | University of Maryland, Baltimore County |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 28°C |
| GPS Coordinates | 9.58 N, 76.52 E Map |
| Discovery Notes | Isolated from a donated soil sample collected in Kottayam, India on 3/20/16. |
| Naming Notes | Scap1, because this will be, we believe, the first sequenced S. scabiei phage. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Dec 13, 2016 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina Sequencing |
| Genome length (bp) | 43060 |
| Character of genome ends | Unknown |
| Overhang Length | 9 bases |
| Overhang Sequence | CGCCGCCCT |
| GC Content | 60.9% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | BI |
| Subcluster | BI2 |
| Cluster Life Cycle | Lytic |
| Other Cluster Members |
|
| Annotating Institution | Unknown or unassigned |
| Annotation Status | In GenBank |
| Plaque Notes | Approximately 1 mm when grown on S. scabiei 36 - 48 hours on NA at 28 deg. C. |
| Morphotype | Siphoviridae |
| Number of Genes | 55 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Annotation in DNA Master Format | Download |
| Submitted Minimal DNA Master File | Download |
| Author List | Download |
| Annotation Cover Sheet | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MF975637 |
| Refseq Number | None yet |
| Published in Paper | Blocker D et al 2018, Genome Announcements: Complete Genome Sequences of Six BI Cluster Streptomyces Bacteriophages, HotFries, Moozy, Rainydai, RavenPuff, Scap1, and SenditCS. |
| Archiving Info |
| Archiving status |
Archived |
| Pitt Freezer Box# |
35 |
| Pitt Freezer Box Grid# |
B1 |
| Available Files |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| Virtual Digest Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |