Mycobacterium phage WhyZach
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage WhyZach | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Josh and Ethan |
| Year Found | 2017 |
| Location Found | Spokane, WA USA |
| Finding Institution | Gonzaga University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 22°C |
| GPS Coordinates | 47.667784 N, 117.403439 W Map |
| Naming Notes | This phage is named after Gonzaga basketball player Zach Collins. He could potentially be leaving Gonzaga if he gets drafted for the NBA this year, but we really don't want him to do that, so WhyZach? |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Lysogeny Notes | When examining the restriction enzymes that reacted with our phage DNA via gel electrophoresis, the only restriction enzyme to cut the phage's DNA was SalI. SalI only produced cuts in the DNA at approximately 6,000 base pairs and below. There was a high concentration of phage DNA in the samples with the following restriction enzymes: ClaI, EcoRI, HindIII, and SalI. BamHI and HaeIII had a lower concentration of phage DNA due to them having darker columns in the gel. |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
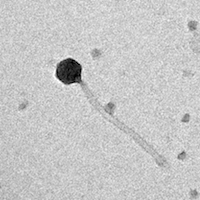
| Plaque Notes | oWhen looking at our TEM image, Our phage has a small head (50 nm) and a long tail (220.83 nm), totaling 270.83 nm. It appears to be part of the siphoviridae family due to its long tail. |
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Pitt Freezer Box# | 65 |
| Pitt Freezer Box Grid# | G6 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |