Mycobacterium phage Beautiphul
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Beautiphul | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Leda Solaimani |
| Year Found | 2012 |
| Location Found | Pullman, WA USA |
| Finding Institution | Washington State University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | Not entered |
| GPS Coordinates | 46.729711 N, 117.173408 W Map |
| Discovery Notes | Discovered from about half an inch below soil surface about a foot from a river. Soil very damp. Few feet away from trees and weeds and a couple plants. |
| Naming Notes | This phage has a very diverse morphology which I think makes it unique and in turn beautiful.I originally wanted to name it Beautiphage, but when I learned that was not possible the name became Beautiphul. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
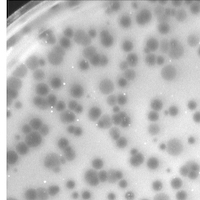
| Plaque Notes | The plaques range from about 0.1-0.35mm in size, and contain bulls eyes,comet tails, and halos. Comet tails are more often larger than smaller but both of these characteristics exist. |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| SEA Designator | 2012WASUbeautiphulLVS |
| SEA Lysate Titer | 4.5 x 10^9 |
| Date of SEA Lysate Titering | Nov 8, 2012 |
| Pitt Freezer Box# | 42 |
| Pitt Freezer Box Grid# | 34 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
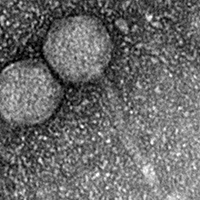
| EM Picture | Download |