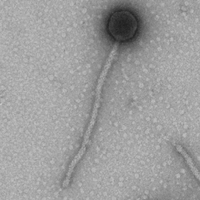
Mycobacterium phage DreamWeaver
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage DreamWeaver | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Former names | TreeHugger2016 |
| Found By | Hayley Nicole Miller |
| Year Found | 2016 |
| Location Found | Bowling green, KY USA |
| Finding Institution | Western Kentucky University |
| Program | Kentucky Biomedical Infrastructure Network Small Genomes Discovery Program |
| From enriched soil sample? | Yes |
| Isolation Temperature | 30°C |
| GPS Coordinates | 36.977253 N, 86.475939 W Map |
| Discovery Notes | The soil sample this phage came from was retrieved from a slightly damp, cool, shady area. The soil obtained was right up against what is believed to be a Kwanzan cherry tree, and retrieved from a depth of four centimeters under the surface. |
| Naming Notes | It was named DreamWeaver due to the tails of the phage having a braided like structure as if it was weaved, and the weaved look helped me come up with DreamWeaver. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Lysogeny Notes | plaques are mostly turbid. but when doing a spot test, the spots are turbid with a few clear plaques here and there. |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
| Plaque Notes | plaques are each about one millimeter in size. They're circular and turbid. |
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| SEA Designator | 2016WEKUdreamweaverHNM |
| SEA Lysate Titer | 2.5 x 10^10 pfu/mL |
| Date of SEA Lysate Titering | Oct 14, 2016 |
| Pitt Freezer Box# | 37 |
| Pitt Freezer Box Grid# | B10 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |