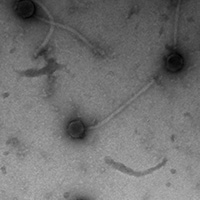
Mycobacterium phage Skip
Add or modify phage thumbnail images to appear at the top of this page.
Duplicate of Marvin
After sequencing by Ion Torrent and assembly, the genome matched Marvin's 100%. Though this is a phage isolated by a high school student working at the University of Pittsburgh, and Marvin was found by Cabrini College, we did have Marvin in our labs at Pitt. Thus, the assumption is that this was a lab contaminant at Pitt.
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Skip | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Rachael Rush |
| Year Found | 2011 |
| Location Found | Pittsburgh, PA USA |
| Finding Institution | University of Pittsburgh |
| Program | Phage Hunters Integrating Research and Education |
| From enriched soil sample? | No |
| Isolation Temperature | Not entered |
| GPS Coordinates | 40.52103 N, 80.077731 W Map |
| Discovery Notes | Duplicate of Marvin, contamination must have occured at some point during process. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Ion Torrent Sequencing |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Available Files | |
| EM Picture | Download |