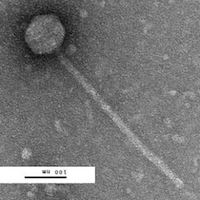
Corynebacterium phage Irvin
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Irvin | |
| Discovery Information | |
| Isolation Host | Corynebacterium xerosis ATCC 373 |
| Found By | Alec Ayala |
| Year Found | 2018 |
| Location Found | Birmingham, AL USA |
| Finding Institution | University of Alabama at Birmingham |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | 37°C |
| GPS Coordinates | 33.543682 N, 86.779633 W Map |
| Discovery Notes | This phage has consistently proven to have an absurd high titer seen with the MVL as well as the HVL. The initial HVL, following 6 Plate Infection, has been calculated to be 1.0*10e16 pfu/mL. The phage may also cause some issues with DNA Extraction due to its high titer. |
| Naming Notes | Named after Honors/AP Biology instructor at Notre Dame High School in Chattanooga, TN |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
| Plaque Notes | Corynebacteriophage Irvin consistently formed pin-pricked sized round plaques following isolation. A set of serial dilutions was prepared up to 10e-13 to provide the image included within this entry. |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Pitt Freezer Box# | 89 |
| Pitt Freezer Box Grid# | C6 |
| Available Files | |
| Plaque Picture | Download |
| EM Picture | Download |