 Phamerator
PhameratorWhat is it?
Phamerator is a comparative genomics and genome exploration tool designed and written by Dr. Steve Cresawn of James Madison University.
In 2017, Phamerator transitioned from a Linux-based program to be a cross-platform web-based program. It is available at https://phamerator.org/
A Phamerator Map
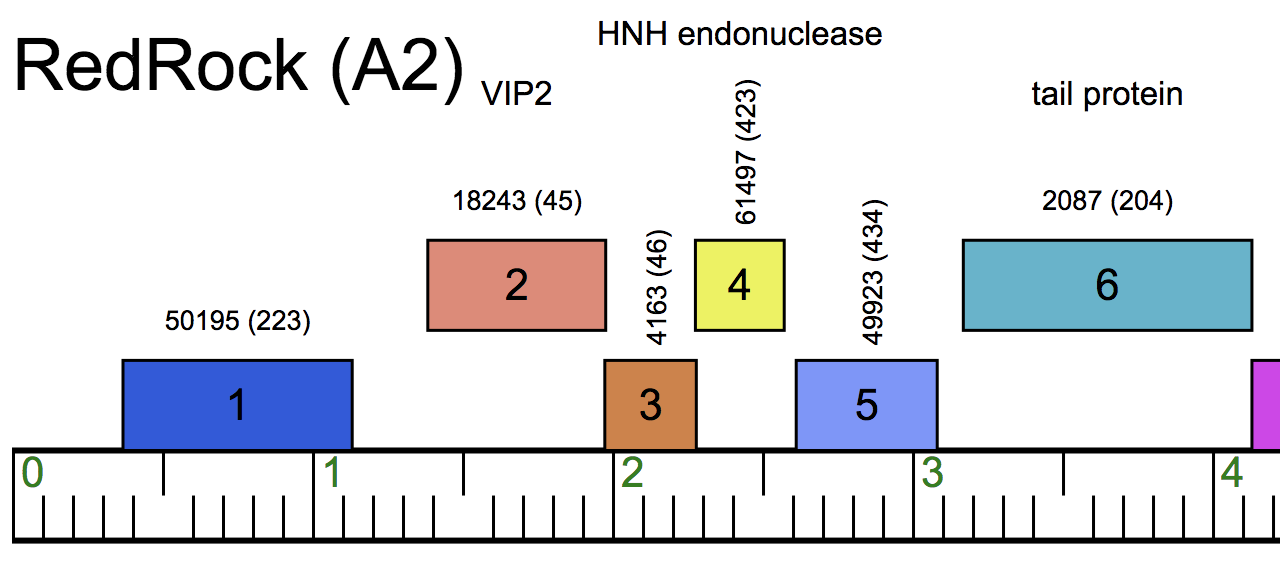
BELOW HERE ARE INSTRUCTIONS FOR THE "LEGACY" VERSION OF PHAMERATOR THAT RUNS ON LINUX. WE RECOMMEND USING PHAMERATOR ON THE WEB RATHER THAN THE LINUX VERSION NOW.
Operating System
Phamerator requires the Ubuntu 12.04 operating system. If you are running Windows or Mac OS X, see the instructions below on how to install an emulator (like Virtual Box) with an Ubuntu virtual machine.
System Requirements
Native Minimum System Requirements
Ubuntu 12.04 "Precise Pangolin"
1 GHz processor
1 GB RAM
128 MB video memory
1 GB free hard-drive space
Internet connection
FULL sudo PRIVILEGES
Emulated Minimum System Requirements (using Ubuntu 12.04 virtual machine)
Windows XP/Vista/7 or at least Mac OS X 10.5
1.6 GHz x86/x64 dual-core processor
2 GB RAM
128 MB video memory
10 GB free hard-drive space
Internet connection
Emulated Minimum System Requirements (using Ubuntu 12.04 virtual machine)
Installation
If you are a SEA-PHAGES member or school, you can get the SEA Virtual Machine which comes with Phamerator pre-installed. It's available here on seaphages.org.
For installation of Phamerator onto a computer already running Ubuntu 12.04, follow these instructions.
For installation of Phamerator onto a computer already running Ubuntu 12.04, follow these instructions.
Getting Started
Once Phamerator is installed, you can follow the instructions in this short Quick Start Guide to start exploring phamerated maps of genomes.
Complete Annotation Guide
Information on how to use Phamerator to assist with annotation of your phage genome can be found in section 6 of the complete DNA Master Annotation Guide.