Mycobacterium phage Relish
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Relish | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Regan Pritchett, Lauren Jaworowski |
| Year Found | 2024 |
| Location Found | Spokane, WA United States of America |
| Finding Institution | Gonzaga University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 22°C |
| GPS Coordinates | 47.6381 N, 117.4211 W Map |
| Naming Notes | When we were brainstorming names, we were very hungry, as our lab time is during the lunch hour. We first thought of a nice peanut butter and jelly sandwich, like an Uncrustable. Mmm. Then, we thought of hotdogs and the different toppings, which gave us the idea of Relish. For our phage, we thought that Relish might be a better name than Uncrustable. The rest is history. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
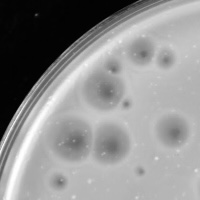
| Plaque Notes | Relish formed a variety of circular plaques that had a clear center and were surrounded by a cloudy halo. Plaques from Relish ranged from 1mm to 10mm throughout the process. The range of plaque sizes for Relish would normally suggest more than one type of phage in the population, however, later experiments showed that it is a single, clonal phage population. |
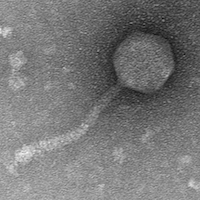
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Not in Pitt Archives |
| Available Files | |
| Plaque Picture | Download |
| EM Picture | Download |