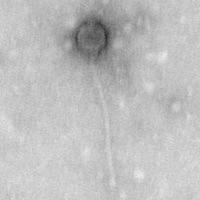
Gordonia phage Algernon
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Algernon | |
| Discovery Information | |
| Isolation Host | Gordonia terrae 3612 |
| Found By | Brianna Kramer |
| Year Found | 2016 |
| Location Found | Pittsburgh, PA USA |
| Finding Institution | University of Pittsburgh |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 30°C |
| GPS Coordinates | 40.444225 N, 79.952492 E Map |
| Discovery Notes | Temp : 13.33ºC Time: 8:16 AM 9/12/16 Description: weather is sunny, slightly chilly, soil is dark, clay-like, underneath mulch, moist; |
| Naming Notes | The name "Algernon" has no special significance, I just enjoy it. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Lysogeny Notes | Algernon is most likely a temperate phage, which explains why the plaques are cloudy. Bacterial host cells that haven't been lysed appear inside a plaque. |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
| Plaque Notes | The plaques are cloudy, round, and uniform morphologies are found. One is large and has hazy edges, the other is pinpoint. Both sized plaques occur as a result of the same phage. |
| Morphotype | Myoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| SEA Lysate Titer | 1.8 X 10^11 |
| Date of SEA Lysate Titering | Oct 12, 2016 |
| Pitt Freezer Box# | 42 |
| Pitt Freezer Box Grid# | B1 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |