Gordonia phage BarkAlicious
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage BarkAlicious | |
| Discovery Information | |
| Isolation Host | Gordonia rubripertincta Grub38 |
| Found By | Rosemary Kinnison and Andrea Breckenridge |
| Year Found | 2025 |
| Location Found | TUCSON, AZ United States |
| Finding Institution | Gonzaga University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 21°C |
| GPS Coordinates | 32.22784 N, 110.92562 W Map |
| Discovery Notes | The soil was very dry and collected from a garden in Tucson, Arizona. |
| Naming Notes | It all started with the final bark of lab partner Andy's dog..... 5 long and dark years ago. Then, she met lab partner Rose, and her life completely changed. Together, their initials echoed this emotionally charged bark. And so, BarkAlicious was born. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
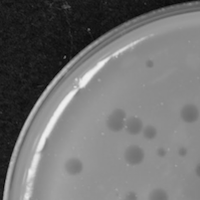
| Plaque Notes | The plaques average a size of 2.6 mm. They are clear, not turbid or cloudy. In the 10^6 dilution, there are approx 83 PFUs. |
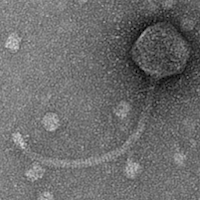
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Not in Pitt Archives |
| SEA Lysate Titer | 3.2x10^10 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |