Gordonia phage Celestia
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Celestia | |
| Discovery Information | |
| Isolation Host | Gordonia terrae 3612 |
| Found By | Seohyun Janice Im |
| Year Found | 2016 |
| Location Found | Pittsburgh, PA USA |
| Finding Institution | University of Pittsburgh |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | 30°C |
| GPS Coordinates | 40.443 N, 79.954 W Map |
| Discovery Notes | The soil sample in which Celestia was found was collected on 9/25/16 at 11:52 PM, the night before lab, in the flowerbed behind the panther statue near the William Pitt Union. The soil sample was collected in a flowerbed containing various kinds of vegetation, including flowers and also other plants resembling lettuce (I dug approximately 2 in. below the surface around a the lettuce-like plant in the flowerbed). Outdoor conditions during soil collection were as follows: the ambient temperature was 62° F and it had not rained the day before soil collection or around the time of soil collection. Characteristics of the soil: the soil was damp and cool. The soil is a blackish color and is not too soft and crumbly but not too hard and chunky either in texture. The soil does not contain many wood chips or chunky bark and seems to contain a lot of organic material. Celestia was found among one of four different morphologies of plaques (small, clear; medium, clear; medium, cloudy; large, cloudy) in the resulting direct isolation plate. After completing the third round of plaque purification, in which I purified two plaques of differing size morphology from the second round of plaque purification, I discovered Celestia on the set of plates containing a small and clear plaque morphology. |
| Naming Notes | With every plate that was held up to the light, the plaque morphology combined with the pattern of plaques on the plate reminded me of the stars and the heavens, thus the name "Celestia". |
| Sequencing Information | |
| Sequencing Complete? | No |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
| Plaque Notes | Celestia creates plaques that are mostly small in terms of size and clear in terms of clarity. Plaque sizes are not always homogeneous-Celestia tends to create plaque sizes that range from small to medium (1-2 mm). |
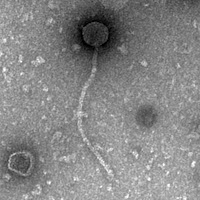
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| SEA Lysate Titer | 1.3 * 10^10 pfu/mL |
| Date of SEA Lysate Titering | Nov 7, 2016 |
| Pitt Freezer Box# | 42 |
| Pitt Freezer Box Grid# | C2 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |