Streptomyces phage ChasesFault
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage ChasesFault | |
| Discovery Information | |
| Isolation Host | Streptomyces mirabilis NRRL B-2400 |
| Former names | ItsChasesFault |
| Found By | Junior Abissi, Angelina Mccreesh |
| Year Found | 2023 |
| Location Found | Catonsville, MD United States |
| Finding Institution | University of Maryland, Baltimore County |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | 30°C |
| GPS Coordinates | 39.252806 N, 76.705145 W Map |
| Naming Notes | We blamed our TA, Chase, for any contamination or failed attempts at the procedures (as a joke). |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Lysogeny Notes | After 2 week incubation time, no apparent mesas formed. This indicates that ChasesFault is an, assumed, lytic/virulent phage. |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
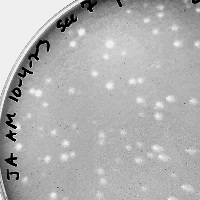
| Plaque Notes | Turbid, small and rough (roughly 0.8-1.0mm). |
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Pitt Freezer Box# | 171 |
| Pitt Freezer Box Grid# | B4 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |