Mycobacterium phage Christoria
Duplicate of Zavala
Upon sequencing, Christoria was found to be 100% identical to Zavala.
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Christoria | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Chris Kropog & Victoria Perez |
| Year Found | 2016 |
| Location Found | Corpus Christi, TX USA |
| Finding Institution | Del Mar College |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | Not entered |
| GPS Coordinates | Unavailable |
| Discovery Notes | Discovered on 8/29/16 at 12:12pm. Found by a tree in front of a building in very dark, moist, and muddy soil near plants. The temperature was 77 degrees Fahrenheit. |
| Naming Notes | The name is a combination of our names. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Approximate Shotgun Coverage | 477 |
| Genome length (bp) | 59969 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 11 bases |
| Overhang Sequence | CTCGTAGGCAT |
| GC Content | 66.6% |
| Fasta file available? | No |
| Characterization | |
| Cluster | K |
| Subcluster | K1 |
| Cluster Life Cycle | Temperate |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
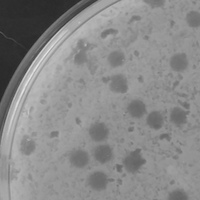
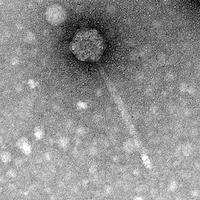
| Plaque Notes | Our phage switched from lysogenic to lytic. The size of our plaque is 5mm in diameter. Phage capsid diameter: 68nm Phage tail length: 221nm |
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| SEA Lysate Titer | 4.7x10^9 |
| Pitt Freezer Box# | 34 |
| Pitt Freezer Box Grid# | G8 |
| Available Files | |
| Plaque Picture | Download |
| EM Picture | Download |