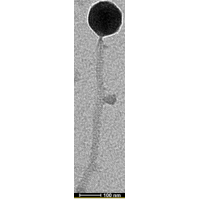
Mycobacterium phage Cram
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Cram | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Clarissa A Martinez, Macie R Skinner, Robel Tadele |
| Year Found | 2022 |
| Location Found | Longview, TX United States of America |
| Finding Institution | LeTourneau University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 37°C |
| GPS Coordinates | 32.4646 N, 94.72703 E Map |
| Discovery Notes | Our sample was collected on August 23, 2022, at 10:13 AM CST. The weather was partially sunny, and the ambient temperature was 22.8°C, the high was 28.3°C and the low was 22.8°C, for the day. The humidity was 90%, as it had rained 2.45 inches in the last 24 hours. The sample was collected near the back entrance of the Glaske Center for Science, Technology, and Engineering building at LeTourneau University in Longview, Texas, USA. The soil was moist and full of organic matter. |
| Naming Notes | We initially used our initials C, R, and M as a placeholder. However, the name stuck when we joked around about how we cram information about our class and phage into our brains. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
| Plaque Notes | The plaques were clear and had an average diameter of 0.83mm (range 0.4-1.0mm, n=10). |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| SEA Lysate Titer | 6.0x10^9 pfu/mL |
| Pitt Freezer Box# | 150 |
| Pitt Freezer Box Grid# | D7 |
| Available Files | |
| Plaque Picture | Download |
| EM Picture | Download |