Mycobacterium phage Fiasco
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Fiasco | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Sara Coppins, Daniela Tejada |
| Year Found | 2025 |
| Location Found | Columbia Falls, MT United States |
| Finding Institution | Gonzaga University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 25°C |
| GPS Coordinates | 48.395288 N, 114.205481 W Map |
| Naming Notes | Found it outside a hotel in Columbia Falls, experimentation caused a fiasco. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
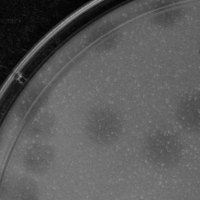
| Plaque Notes | We used our 10^-6 dilution sample resulting in 0.6 cm circular and clear plaques dispersed throughout the plate. Labeling was visible on all sides of the plate, used an image with the clearest visible area. |
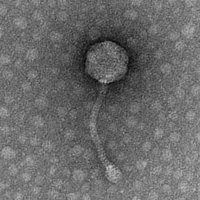
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Not in Pitt Archives |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |