Mycobacterium phage IronFlat
Add or modify phage thumbnail images to appear at the top of this page.
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage IronFlat | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Roxy Witham, Pippa Liddiard |
| Year Found | 2020 |
| Location Found | Boulder, CO United States |
| Finding Institution | University of Colorado Boulder |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | 37°C |
| GPS Coordinates | Unavailable |
| Discovery Notes | We discovered our phage from an isolated soil sample. This soil sample was found in a moist patch of dirt that was covered with snow under a tree outside the Porter Bioscience building. |
| Naming Notes | We decided to name our phage after the Flatiron mountains in Boulder, Colorado which is where our soil sample originated from. Due to the fact that Flatiron was already taken, we chose to name our phage IronFlat instead. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
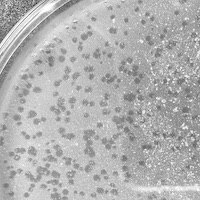
| Plaque Notes | Our plaques identified were clear, circular plaques suggesting the the phage we isolated was lytic. The plaques formed on our bacterial lawn were roughly 1-2mm in diameter. |
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Pitt Freezer Box# | 126 |
| Pitt Freezer Box Grid# | G9 |
| Available Files | |
| Plaque Picture | Download |
| EM Picture | Download |