Mycobacterium phage Lafarge
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Lafarge | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Nathan Frey |
| Year Found | 2025 |
| Location Found | Philadelphia, PA United States |
| Finding Institution | Saint Joseph's University |
| Program | Unavailable |
| From enriched soil sample? | No |
| Isolation Temperature | 37°C |
| GPS Coordinates | 39.992816 N, 75.23981 W Map |
| Discovery Notes | A soil sample was collected from the base of a tree located between Sourin and Lafarge at Saint Joseph's University. At the time of collection, the weather was sunny and humid, though the soil itself was dry. Direct isolation of the sample revealed the presence of multiple bacteriophages, and this finding was subsequently confirmed through three rounds of purification. After the Electron-Micrograph imaging, the average tail length was 89.5 nm. The average capsid size was 49.3 nm. A total of three phage particles were measured. |
| Naming Notes | During the early arrival program for Beagle students, we spent three days in LaFarge, an old and worn-down building, while waiting for the new dormitory to be completed. Despite its age and slightly eerie charm, it turned out to be a very productive location: soil collected from the building yielded multiple phages. In honor of the unique character of LaFarge and the fact that the phages were first discovered there, the phage was named after the building. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
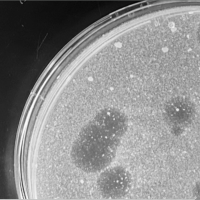
| Plaque Notes | The phage plaques have a size of 4-5 mm long in diameter. The plaques are clear. Each phage is surrounded by a halo that is 0.5-1 mm in diameter. The halo is slightly cloudy. 86 were measured. Phage were incubated 48 hours. |
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Not in Pitt Archives |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |