Mycobacterium phage Mathis22
Add or modify phage thumbnail images to appear at the top of this page.
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Mathis22 | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Mason Mathis |
| Year Found | 2022 |
| Location Found | Glasgow, KY United States |
| Finding Institution | Western Kentucky University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 30°C |
| GPS Coordinates | 36.99392 N, 85.89769 W Map |
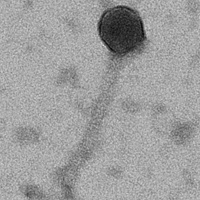
| Discovery Notes | Mathis22 was found in an enriched soil sample from Gorin park in Glasgow, KY. The first step of discovery was spot plating where I obtained turbid bullseye plaques 15mm in size. Through a series of four dilution series, the phage was isolated until the dilutions were 10-fold and the plaque morphology and size remained the same. Only once, during dilution 3, were any of the plaques not turbid bullseye. A few of the plaques were clear. The size of the plaques remained between 2.5mm and 7.5mm. A one plate lysate was then obtained with a titer of 2.01E11. A ten plate lysate was next with a high titer of 2.8E10. Using the high titer lysate, pictures were taken of the phage with an electron microscope. The average tail length of Mathis22 is 147.396nm and the average capsid diameter is 48.090nm. The next steps were isolating and purifying the phage DNA. The phage DNA was exposed through the process of incubating with nuclease mix and then mixing with phage precipitate solution. The DNA was then isolated by first washing with DNA clean-up resin and then two washes of 80% isopropanol to remove proteins and salt from the DNA. The DNA was then eluted twice with elution buffer to obtain clean DNA. My DNA concentration was 13.1ng/uL, which was too low, so I repeated the process of DNA isolation and purification for a new concentration of 50.3ng/uL. The final steps were to run a restriction enzyme digest. 10uL of the 50.3ng/uL DNA was added to the enzymes BamHI, ClaI, EcoRI, HaeIII, and HindIII. A gel showed no cuts were made in the phage genome by BamHI, EcoRI, or HindIII. ClaI made cuts at 10,000bp, 5,000bp, and 4,000bp. HaeIII had a plethora of cuts that appeared as a smear on the gel. Finally, I repeated that process with the enzymes AhdI, BstXI and DraIII-HF. A gel showed no cuts made by BstXI or DraIII-HF. AhdI had a distinct cut at 6,000bp, multiple cuts between 4,000bp and 3,000bp and a smear from 2,000bp down. |
| Naming Notes | Mathis22 comes from my last name being Mathis and the phage being discovered in 2022. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
| Plaque Notes | The plaques for Mathis22 started as turbid bullseye plaques with defined halos and remained that way. A very few amount of times clear plaques would show up on a plate with turbid bullseye plaques, but they didn't show up often. The majority of the plaques measured about 5mm, but occasionally they would be as small as 2.5mm or as large as 7.5mm. |
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| SEA Lysate Titer | 2.8E10 |
| Pitt Freezer Box# | 157 |
| Pitt Freezer Box Grid# | G7 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |