Mycobacterium phage Nexus
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Nexus | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Daniel Pelyhes |
| Year Found | 2015 |
| Location Found | Augusta, MI USA |
| Finding Institution | Hope College |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | Not entered |
| GPS Coordinates | 42.38 N, 85.38 W Map |
| Discovery Notes | Collected from an animal pasture not far from my house. The area contained donkeys and goats. The sample was taken from a depth of approximately 6 cm in relatively dry soil. |
| Naming Notes | A nexus is a connection between two or more things. My phage morphology is large and at higher concentrations the way they clustered made the bacteria between the plaques look like pathways, hence the name Nexus |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
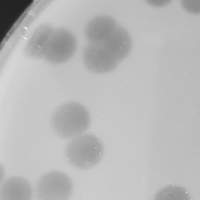
| Plaque Notes | The plaques are on average around 5.8mm. They are large lytic plaques with clear and distinct edges. |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Pitt Freezer Box# | 19 |
| Pitt Freezer Box Grid# | E1 |
| Available Files | |
| Plaque Picture | Download |
| EM Picture | Download |