Mycobacterium phage PeanutandJelly
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage PeanutandJelly | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Payton Jones/Jamie Vulgamore |
| Year Found | 2012 |
| Location Found | Corpus Christi, TX USA |
| Finding Institution | Del Mar College |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | Not entered |
| GPS Coordinates | 27.454435 N, 97.242376 W Map |
| Discovery Notes | The sample came from a wet, dark-colored topsoil grass bed (3 cm deep) in the shade of a tree. The grass bed was flooded with a pipe malfunction underground, that came through the cracks in the concrete and flooded the street and grass bed. The time collected was 11:50 am, henceforth the hottest part of the day has yet to come, the temperature was 99F (37.2C). |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
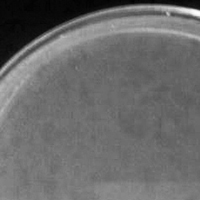
| Plaque Notes | The plaques are, on average, 1 mm in diameter (area: 3.14mm2) and circular in shape. The plaques are produced in the temperate cycle and often very difficult to see. Plaques in the first stages of purification, up unto the Titer Assay, are difficult to see. The Titer Assay took the physical appearance of a lytic cycle phage, however afterwards on the Emperical Testing and onward the phages took the appearance of temperate phage cycle as before. |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Pitt Freezer Box# | 35 |
| Pitt Freezer Box Grid# | 27 |
| Available Files | |
| Plaque Picture | Download |
| EM Picture | Download |