Mycobacterium phage Phipolypsi
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Phipolypsi | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Gabriel Page |
| Year Found | 2019 |
| Location Found | Blacksburg, VA United States |
| Finding Institution | Virginia Tech |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 37°C |
| GPS Coordinates | 37.223889 N, 80.423611 W Map |
| Discovery Notes | Phage acquired from a soil sample taken at 1:00pm outside of Fralin Hall on 8/29/2019. Outside temperature of 29C and low humidity. The extracted soil was 7cm deep from the surface. |
| Naming Notes | Phipolypsi was selected as the name of my bacteriophage because of my dedication to the Phi Kappa Psi fraternity and Virginia Polytechnic Institute and State University where I attend. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
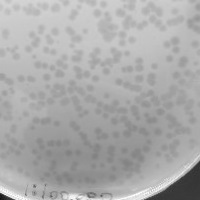
| Plaque Notes | The phage plaques were lytic with large clearings. This was found evident in every experiment. |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Pitt Freezer Box# | 102 |
| Pitt Freezer Box Grid# | C1 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
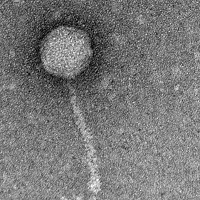
| EM Picture | Download |