Mycobacterium phage Raspberry
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Raspberry | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Megha Talur |
| Year Found | 2014 |
| Location Found | Baltimore, MD USA |
| Finding Institution | Johns Hopkins University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | Not entered |
| GPS Coordinates | 39.32817 N, 76.620798 W Map |
| Discovery Notes | Original soil sample was collected from a sunflower patch on the Johns Hopkins University Homewood campus at a depth of 2.6 cm. The soil sample was dry and obtained on August 27, 2014 at 3:07 pm and 88 degrees Fahrenheit. |
| Naming Notes | The name for this phage came from my favorite fruit and color-raspberry! Additionally, as I got farther into the isolation my plaque morphology became smaller and smaller reminding me of raspberry seeds. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
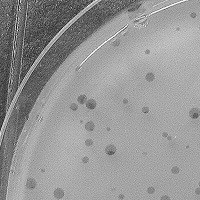
| Plaque Notes | Original morphology of plaque was medium sized, round, and clear. As isolation progressed the morphology shifted to a slightly smaller size but still round and clear. Thus, the final morphology is a small-medium, round, and clear plaque. |
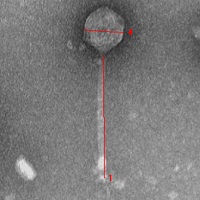
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| SEA Lysate Titer | 7.5x10^(9) |
| Date of SEA Lysate Titering | Nov 12, 2014 |
| Pitt Freezer Box# | 14 |
| Pitt Freezer Box Grid# | C2 |
| Available Files | |
| Plaque Picture | Download |
| EM Picture | Download |