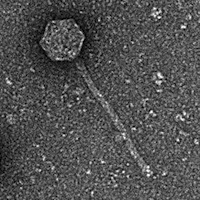
Mycobacterium phage SCFlamingo
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage SCFlamingo | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Keqing Li |
| Year Found | 2017 |
| Location Found | Santa Cruz, CA USA |
| Finding Institution | University of California, Santa Cruz |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | Not entered |
| GPS Coordinates | Unavailable |
| Discovery Notes | The environmental soil sample was collected from UCSC Life lab compost pile. |
| Naming Notes | I decide to name the unknown phage “flamingo” due to the presence of end plate and the body of the phage, which resembles the appearance of a flamingo. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Sequencing Facility | University of California, Santa Cruz |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Singleton |
| Subcluster | -- |
| Cluster Life Cycle | Unknown |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
| Plaque Notes | After 5 times of passage, figure 2 shows the semi-transparent small plaques, indicating that the phage might be lysogenic. The semi-transparent morphology indicates that M.smegmatis is still alive within the plaques. |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Not in Pitt Archives |
| SEA Lysate Titer | 126 ul of 10^-2 |
| Date of SEA Lysate Titering | Oct 5, 2017 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |