Mycobacterium phage Scamp17
Add or modify phage thumbnail images to appear at the top of this page.
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Scamp17 | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Trenton C Sparks |
| Year Found | 2017 |
| Location Found | Irvine, KY USA |
| Finding Institution | Eastern Kentucky University |
| Program | Kentucky Biomedical Infrastructure Network Small Genomes Discovery Program |
| From enriched soil sample? | No |
| Isolation Temperature | Not entered |
| GPS Coordinates | 37.782778 N, 84.157222 W Map |
| Discovery Notes | Dirt sample was taken from soil next to creek off the road. Soil was moist but not wet. Sample was taken to the lab within 24 hours and started isolation. |
| Naming Notes | Soil was retrieved next to Station Camp Creek in Irvine, Ky. This is where Scamp comes from. The S from station and then camp following. 17 Comes from the year found, 2017. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
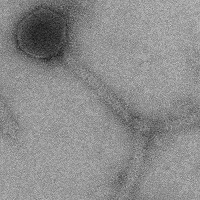
| Plaque Notes | Phage was a typical bacteriophage with a 51.8nm head and a 189.6nm tail with a brush on the end of the tail. |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Not in Pitt Archives |
| Available Files | |
| EM Picture | Download |