Microbacterium phage Sepiida
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Sepiida | |
| Discovery Information | |
| Isolation Host | Microbacterium foliorum NRRL B-24224 |
| Former names | BadLabmate |
| Found By | Kavan Jackson |
| Year Found | 2022 |
| Location Found | Oxford, OH United States |
| Finding Institution | Miami University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 30°C |
| GPS Coordinates | 39.741667 N, 84.573889 W Map |
| Discovery Notes | Sample Came from pine needle litter. |
| Naming Notes | The name came from the inconsistent nature of the phage. It randomly decides to become more or less lytic making it hard to find when needed. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Sequencing Facility | University of Pittsburgh Genomics and Proteomics Core Laboratories |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | EH |
| Subcluster | -- |
| Cluster Life Cycle | Temperate |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
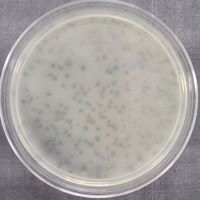
| Plaque Notes | The plaque was a small, cloudy circle with consistent edges. |
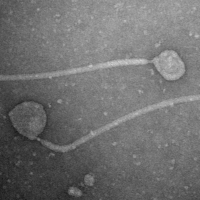
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| SEA Lysate Titer | 5.0*10^8 |
| Pitt Freezer Box# | 160 |
| Pitt Freezer Box Grid# | A10 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |