Mycobacterium phage Tafa
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Tafa | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Ashley Tafadzwa Nerwande |
| Year Found | 2017 |
| Location Found | Durban, South Africa |
| Finding Institution | University of Kwazulu-Natal |
| Program | Mycobacterial Genetics Course, Durban, South Africa |
| From enriched soil sample? | Unknown |
| Isolation Temperature | Not entered |
| GPS Coordinates | 29.48593 S, 30.56348 E Map |
| Discovery Notes | Soil was sampled from upper rich soil (2 cm deep) The soil sample was taken from an area on University of KwaZulu Natal Westville Campus( F-BLOCK near the entrance of Discipline of Clinical Anatomy),near a shrub plant. The area was relatively wet and the sample was then enriched |
| Naming Notes | The name was derived from my second name “Tafadzwa”, which means we are pleased in Shona. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
| Plaque Notes | The plaques were large, round and clear. |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Not in Pitt Archives |
| SEA Lysate Titer | 9x10^12 |
| Available Files | |
| Plaque Picture | Download |
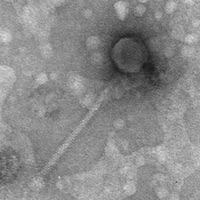
| EM Picture | Download |