Mycobacterium phage TopPatt
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage TopPatt | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Caitlyn Patterson |
| Year Found | 2023 |
| Location Found | Glasgow, KY United States of America |
| Finding Institution | Western Kentucky University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 30°C |
| GPS Coordinates | 37.0175 N, 85.966389 W Map |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Lysogeny Notes | During spot plating protocol, a mesa formation was present in the 10^0 clearing when incubated at 37℃. This formation is evidence that Mycobacterium smegmatis is being induced as a lysogen. This spot formation was used to produce a T-Streak plate. A colony from this T-Streak plate was inoculated at 37℃ until the liquid media was turbid. This inoculation was used to set up a second set of spot plates. The lysogen plated at 37℃ showed no growth which verified that a lysogen was isolated. The lysogen plated 30℃ presented with plaques, indicating spontaneous conversion into the lytic cycle. |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
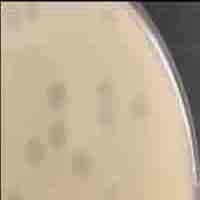
| Plaque Notes | Halos present in plaques |
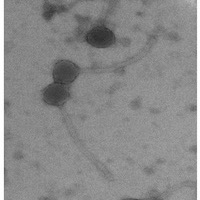
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Not in Pitt Archives |
| SEA Lysate Titer | 9.5 X 10^8 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |