Gordonia phage Wassil
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Wassil | |
| Discovery Information | |
| Isolation Host | Gordonia terrae 3612 |
| Found By | Isabella Sedor |
| Year Found | 2017 |
| Location Found | Pittsburgh, PA USA |
| Finding Institution | University of Pittsburgh |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | 19°C |
| GPS Coordinates | 40.4375 N, 79.9512 W Map |
| Discovery Notes | This phage was discovered in a dirt sample collected close to the University of Pittsburgh, right near Oakland Square, Pittsburgh, Pennsylvania to be more precise. The soil was very dry, but did not have many rocks or sticks in it. The sample was collected at 6:45 p.m. while it was still light and partially cloudy. There were a few trees and bushes near the location as well. The standard direct isolation procedure was used to isolate the phage. After getting results back from this procedure, two plates were used to conduct a spot test. Only one out of the eight spots between both plates was completely cleared. This meant that a phage had finally been found from the evidence that it had infected and killed the bacterial cells in that area. |
| Naming Notes | This name was actually the name of the researcher's grandfather who passed away a few years ago. Not only did she find it to be an interesting name, but because this is such an educational lab full of discovery she figured it would be a good idea to name her phage after someone who valued those things. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Lysogeny Notes | This phage's lysogeny, or whether it is a lytic or temperate phage, was hard to observe at first through direct isolation and spot testing. There was a lot of contamination in both; clear and turbid plaques could be observed in the plates. By carrying forward from the spot test by only using the one completely cleared spot with no contamination, the researcher was able to get definitive results of what type of phage this is. It is a lytic phage. This is certain because all of the many following plaque purifications had the basic same results. The plaques were the same size of roughly 1mm and completely clear. A temperate phage can choose between lytic growth and lysogeny, but a lytic phage only does lytic growth. Because lytic growth causes viral production and cell lysis that creates a clearing, it became obvious after several rounds that this is a lytic phage. |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
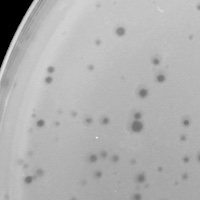
| Plaque Notes | The plaque morphology picture used in this page represents well-spaced plaques. There are about 100 clearings on this plate otherwise covered in an orange color indicating host bacterial cell growth. These plaques are approximately 1 mm in diameter. Something important to note is that the round shape, size, and amount of plaques in this plate follow the pattern of morphology that has occurred through all previous plaque purifications. This is also true for the other plates including cleared plates, webbed plates, and plates with few to no plaques at all. This is a good indication that only one type of phage is being worked with and only one strand of DNA would be sequenced. |
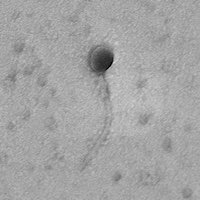
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| SEA Lysate Titer | 3.3x10^10 |
| Pitt Freezer Box# | 58 |
| Pitt Freezer Box Grid# | C1 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |