| Detailed Information for Phage Xkcd426 | |
| Discovery Information | |
| Isolation Host | Streptomyces griseus ATCC 10137 |
| Found By | Preston McDonald, Rajni Majhi |
| Year Found | 2012 |
| Location Found | Valley View, TX USA |
| Finding Institution | University of North Texas |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | Not entered |
| GPS Coordinates | 33.43543 N, 97.21993 W Map |
| Discovery Notes | Discovered during the University of North Texas Transitions Summer Workshop. |
| Sequencing Information | |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Nov 5, 2014 |
| Sequencing Facility | University of North Texas |
| Shotgun Sequencing Method | Ion Torrent |
| Approximate Shotgun Coverage | 186 |
| Genome length (bp) | 64477 |
| Character of genome ends | Circularly Permuted |
| End Determination Notes | No read buildups in shotgun sequencing data. |
| GC Content | 68.8% |
| Sequencing Notes | There were a couple of regions that are probably correct but may have sequencing errors. The first is about around 19370 bp, and the other is around 34920 bp. |
| Fasta file available? | Yes: Download fasta file |
| Characterization | |
| Cluster | BG |
| Subcluster | -- |
| Cluster Life Cycle | Lytic |
| Other Cluster Members |
Click to View |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | In GenBank |
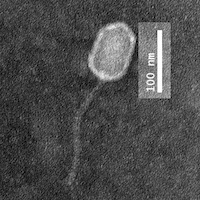
| Morphotype | Siphoviridae |
| Number of Genes | 78 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
Click to ViewXkcd426_1 Xkcd426_2 Xkcd426_3 Xkcd426_4 Xkcd426_5 Xkcd426_6 Xkcd426_7 Xkcd426_8 Xkcd426_9 Xkcd426_10 Xkcd426_11 Xkcd426_12 Xkcd426_13 Xkcd426_14 Xkcd426_15 Xkcd426_16 Xkcd426_17 Xkcd426_18 Xkcd426_19 Xkcd426_20 Xkcd426_21 Xkcd426_22 Xkcd426_23 Xkcd426_24 Xkcd426_25 Xkcd426_26 Xkcd426_27 Xkcd426_28 Xkcd426_29 Xkcd426_30 Xkcd426_31 Xkcd426_32 Xkcd426_33 Xkcd426_34 Xkcd426_35 Xkcd426_36 Xkcd426_37 Xkcd426_38 Xkcd426_39 Xkcd426_40 Xkcd426_41 Xkcd426_42 Xkcd426_43 Xkcd426_44 Xkcd426_45 Xkcd426_46 Xkcd426_47 Xkcd426_48 Xkcd426_49 Xkcd426_50 Xkcd426_51 Xkcd426_52 Xkcd426_53 Xkcd426_54 Xkcd426_55 Xkcd426_56 Xkcd426_57 Xkcd426_58 Xkcd426_59 Xkcd426_60 Xkcd426_61 Xkcd426_62 Xkcd426_63 Xkcd426_64 Xkcd426_65 Xkcd426_66 Xkcd426_67 Xkcd426_68 Xkcd426_69 Xkcd426_70 Xkcd426_71 Xkcd426_72 Xkcd426_73 Xkcd426_74 Xkcd426_75 Xkcd426_76 Xkcd426_77 Xkcd426_78 |
| Submitted Annotation in DNA Master Format | Download |
| Submitted Minimal DNA Master File | Download |
| Publication Info | |
| Uploaded to GenBank? | Yes |
| GenBank Accession | KU530220 |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Pitt Freezer Box# | 124 |
| Pitt Freezer Box Grid# | F12 |
| Available Files | |
| Plaque Picture | Download |
| EM Picture | Download |