Mycobacterium phage ChiT
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage ChiT | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Megan Garcia |
| Year Found | 2016 |
| Location Found | Spokane, WA USA |
| Finding Institution | Gonzaga University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | Not entered |
| GPS Coordinates | 47.67 N, 117.4 W Map |
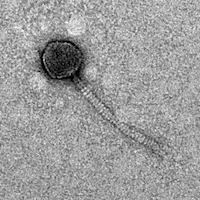
| Discovery Notes | My phage is similar to the phage ByePhelicia. Our phage DNA are cut from the same enzymes (EcoRI, HaeIII, and SalI), but in different sizes of fragments. My phage individually from the TEM pictures are 206nm long. They also do not affect at room temperature. |
| Naming Notes | My phage name is inspired by my Chihuahua, Titan Taco Burrito Garcia, who is back at home in Hawai'i. It is also inspired by my favorite drinks, a hot chai tea and chai tea lattes. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
| Plaque Notes | My phage are fairly random in size (even after many dilutions from one plaque). They are about sizes between 1mm to 3mm in size (making the size not very uniform). They are also clear plaque with a very round shape. |
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| SEA Lysate Titer | 6.2*10^8 pfu/mL |
| Date of SEA Lysate Titering | Jun 14, 2016 |
| Pitt Freezer Box# | 40 |
| Pitt Freezer Box Grid# | D2 |
| Available Files | |
| Plaque Picture | Download |
| EM Picture | Download |