Mycobacterium phage Cumberland
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Cumberland | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Avery Robinson |
| Year Found | 2024 |
| Location Found | Russel Springs, KY United States of America |
| Finding Institution | Western Kentucky University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 30°C |
| GPS Coordinates | 37.01167 N, 84.93144 E Map |
| Discovery Notes | The site is near the perimeter of a house, in an area surrounded by trees. The area sees ample amounts of rain. The site receives low amounts of foot traffic. The soil was damp the day of collection due to adequate rain the night before collection. The area drains downhill in Lake Cumberland. |
| Naming Notes | The name came from the location where the phage was collected from. The property from which the site was collected is near Lake Cumberland. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Lysogeny Notes | The lysogen test suggests that the streaked colonies are lysogens due to the resistance of reinfection by the phage. The phage Cumberland shows lysogeny of the host, Mycobacterium smegmatis. |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
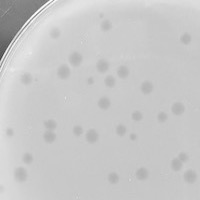
| Plaque Notes | 4.5 mm turbid plaques that appear in circular shapes. |
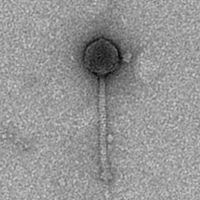
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Not in Pitt Archives |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| Virtual Digest Picture | Download |
| EM Picture | Download |