Mycobacterium phage CurlyFry
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage CurlyFry | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Nathalia Meza and Hanna Lane |
| Year Found | 2024 |
| Location Found | Golden, CO United States |
| Finding Institution | Gonzaga University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 22°C |
| GPS Coordinates | 39.80564 N, 105.21711 W Map |
| Discovery Notes | The dirt sample was collected from Colorado where the weather was cold and dry. There was little to no moisture in the air due to the location being in the desert. |
| Naming Notes | We had chosen our phage name as we found it to be unique. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
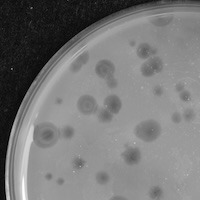
| Plaque Notes | In analyzing Curly Fry's plaque morphology we see the presence of phages through clearings scattered on the lawn. All the clearings present vary in size as some are smaller than others but averaging at about 1 mm for most. Selected phages present on the plate also have transparent centers with the formation of lawn rings around them. |
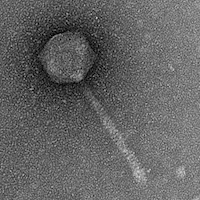
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| SEA Lysate Titer | 3.3 x 10^7 |
| Pitt Freezer Box# | 192 |
| Pitt Freezer Box Grid# | D7 |
| Available Files | |
| Plaque Picture | Download |
| EM Picture | Download |