Microbacterium phage DaeSlime
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage DaeSlime | |
| Discovery Information | |
| Isolation Host | Microbacterium foliorum NRRL B-24224 |
| Found By | David smith, Karmen Whetzel |
| Year Found | 2024 |
| Location Found | Cumberland, MD United States |
| Finding Institution | Allegany College of Maryland |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 29°C |
| GPS Coordinates | 39.653192 N, 78.728943 W Map |
| Discovery Notes | Discovered by David Smith and Karmen Whetzel in the fall of 2024, collected along the river bank of evils creek. Soil type was wet and sandy and temperature was about 20.0 degrees celsius when collected. |
| Naming Notes | Name was made up by David and Karmen was in agreegence, it's a mix of my nickname (dae dae) and Slime a moist, soft, slippery substance. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Sequencing Facility | University of Pittsburgh Genomics and Proteomics Core Laboratories |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
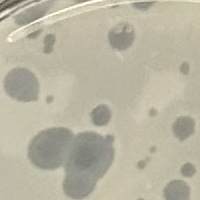
| Plaque Notes | Appeared Small, PFU measured to be 4-5 mm in size, the plaque looks to be clear and had some halo like plaques form early.As we worked it up it was clear our PFU appeared to be clear. |
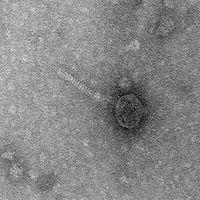
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| SEA Lysate Titer | 2.23 x 10^9 pfu/ml |
| Pitt Freezer Box# | 196 |
| Pitt Freezer Box Grid# | C6 |
| Available Files | |
| Plaque Picture | Download |
| EM Picture | Download |