Mycobacterium phage DarkChelan
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage DarkChelan | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Katherine Duffy and Lauren Buchberger |
| Year Found | 2016 |
| Location Found | Chelan, WA USA |
| Finding Institution | Gonzaga University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 22°C |
| GPS Coordinates | Unavailable |
| Discovery Notes | Found under a few inches (2-4in.) of wet rocks in the mountains of Chelan. The fround was cold and hard from recent snow flurries, however there was no snow on the area where the soil was found. |
| Naming Notes | The name was created using the name of the area in which the sample was found and descriptors of the appearance of the sample before enrichment. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Lysogeny Notes | Our phage grew on both a the host Samuel Plaqueson and at 37 degrees C. From this, we know that as our phage was able to infect Samuel Plaqueson, a K-cluster phage, our phage does not belng to this cluster. Also, when plated on a the M. smegmatis host at 37 degrees C, all of our dilutions of phage were able to grow plaques. In all of this we used the phage "Gumby" as a positive control, which allows us to determine if our phage samples were corectly created if both the samples and Gumby phage grew on the host bacteria. In our case, our phage dilutions were correctly made as we observed growth of plaques by all the phage. |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
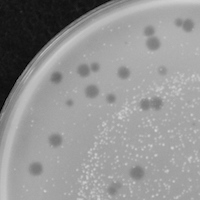
| Plaque Notes | Our plaques are consistently about 1mm in diameter and are clear. This suggests we isolated a lytic phage in whihc the colonies of phage digested away the M.smegmatis host quickly and efficiently. Overall, our plaques are evenly spaced at a high concentration of similar appearances, meaning our phage has been well isolated. |
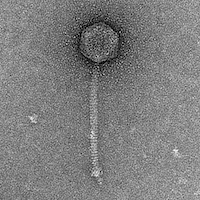
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| SEA Lysate Titer | 1.0X10^10pfu/ml |
| Date of SEA Lysate Titering | Mar 22, 2016 |
| Pitt Freezer Box# | 40 |
| Pitt Freezer Box Grid# | D11 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |