Mycobacterium phage Flip
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Flip | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Samantha Sarli |
| Year Found | 2014 |
| Location Found | Easton, PA USA |
| Finding Institution | Lehigh University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | Not entered |
| GPS Coordinates | 40.727857 N, 75.247505 W Map |
| Discovery Notes | Soil sample obtained on January 11, 2014. Approximate temperature of that day was 41F. The soil was clay-like, damp, and muddy. |
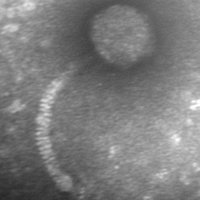
| Naming Notes | The name "Flip" was chosen based on electron microscopy images in which the tails of the phages were bent and very flexible, almost giving the impression that the phage was flipping. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
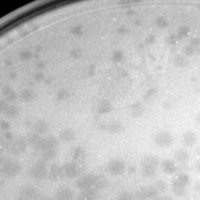
| Plaque Notes | The plaques were consistently turbid and ranged from 0.5mm-3.0mm in diameter. |
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| SEA Lysate Titer | 2.09x10^8 |
| Date of SEA Lysate Titering | Apr 22, 2014 |
| Pitt Freezer Box# | 7 |
| Pitt Freezer Box Grid# | A5 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |