Mycobacterium phage IronZag
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage IronZag | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Dashiell Horstman and Jack Oliver |
| Year Found | 2024 |
| Location Found | Spokane, WA United States |
| Finding Institution | Gonzaga University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 22°C |
| GPS Coordinates | 47.6648 N, 117.4007 E Map |
| Discovery Notes | The sample was taken in the fall on a sunny day with no precipitation. It was taken where landscaping was done, and there were some traces of beauty bark. After we took the sample, we noticed a dead bird inches away. |
| Naming Notes | We named the Phage IronZag after our school. Zag comes from Gonzaga, and Iron is a synonym for strong. So when put together, it means Strong Zag or Strong Students. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
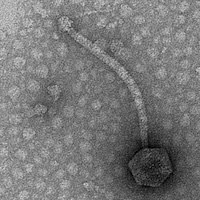
| Plaque Notes | They are tiny, circular, with diffused edges. Their size makes it hard to tell if they are turbid, clear, or haloed. The TEM shows it is a Siphoviridae. |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Pitt Freezer Box# | 192 |
| Pitt Freezer Box Grid# | G6 |
| Available Files | |
| Plaque Picture | Download |
| EM Picture | Download |