Microbacterium phage Labella
Add or modify phage thumbnail images to appear at the top of this page.
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Labella | |
| Discovery Information | |
| Isolation Host | Microbacterium foliorum NRRL B-24224 |
| Found By | Asmaa Abdullah, Deeptha Posa |
| Year Found | 2025 |
| Location Found | Springfield, PA United States |
| Finding Institution | Drexel University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | 30°C |
| GPS Coordinates | 39.91741 N, 75.33815 W Map |
| Discovery Notes | It was found in moist soil with a mixture of mulch on the right side of a rose plant. |
| Naming Notes | Labella was named after the vibe of the color pink, with the addition of science. |
| Sequencing Information | |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Jan 19, 2026 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina NextSeq 1000 |
| Read Type | Single-end reads |
| Read Length | 100 bp |
| Approximate Shotgun Coverage | 972 |
| Genome length (bp) | 41864 |
| Character of genome ends | Circularly Permuted |
| GC Content | 66.9% |
| Fasta file available? | Yes: Download fasta file |
| Characterization | |
| Cluster | EJ |
| Subcluster | -- |
| Cluster Life Cycle | Unknown |
| Other Cluster Members | |
| Annotating Institution | Drexel University |
| Annotation Status | Being Annotated (Expected completion by 5/1/2026) |
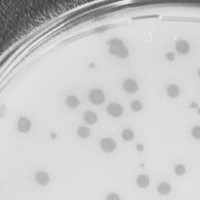
| Plaque Notes | Between 0.3-0.2 mm was the size of the phage after each purification. There were varying sizes of circles within that range. |
| Number of Genes | 61 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
Click to ViewLabella_1 Labella_2 Labella_3 Labella_4 Labella_5 Labella_6 Labella_7 Labella_8 Labella_9 Labella_10 Labella_11 Labella_12 Labella_13 Labella_14 Labella_15 Labella_16 Labella_17 Labella_18 Labella_19 Labella_20 Labella_21 Labella_22 Labella_23 Labella_24 Labella_25 Labella_26 Labella_27 Labella_28 Labella_29 Labella_30 Labella_31 Labella_32 Labella_33 Labella_34 Labella_35 Labella_36 Labella_37 Labella_38 Labella_39 Labella_40 Labella_41 Labella_42 Labella_43 Labella_44 Labella_45 Labella_46 Labella_47 Labella_48 Labella_49 Labella_50 Labella_51 Labella_52 Labella_53 Labella_54 Labella_55 Labella_56 Labella_57 Labella_58 Labella_59 Labella_60 Labella_61 |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Not in Pitt Archives |
| Available Files | |
| Plaque Picture | Download |