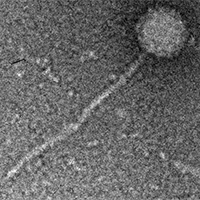
| Detailed Information for Phage Nilo |
| Discovery Information |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Elsie Guevara |
| Year Found | 2012 |
| Location Found | Sheffield, MA USA |
| Finding Institution | Berkshire School |
| Program | Phage Hunters Integrating Research and Education |
| From enriched soil sample? | No |
| Isolation Temperature | Not entered |
| GPS Coordinates | 42.118153 N, 73.413545 W Map |
| Discovery Notes | Collected from an herb garden that had cold, moist, rich soil. |
| Naming Notes | Nilo is derived from my Dad's name, Petronilo. He suggested that I name my first virus after him. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | May 7, 2013 |
| Sequencing Facility | University of Pennsylvania DNA Sequencing Facility |
| Shotgun Sequencing Method | 454 |
| Sequencer Used | 454 Genome Sequencer FLX |
| Approximate Shotgun Coverage | 94 |
| Genome length (bp) | 71752 |
| Character of genome ends | Circularly Permuted |
| GC Content | 56.0% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | R |
| Subcluster | -- |
| Cluster Life Cycle | Lytic |
| Other Cluster Members |
|
| Annotating Institution | University of Pittsburgh |
| Annotation Status | In GenBank |
| Plaque Notes | Nilo makes clear pinprick plaques of about 0.5 mm in diameter, sometimes with uneven edges. |
| Morphotype | Siphoviridae |
| Number of Genes | 102 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Minimal DNA Master File | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MH001447 |
| Refseq Number | None yet |
| Published in Paper | Pope, Bowman, Russell, et al 2015, eLIFE: Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity |
| Archiving Info |
| Archiving status |
Archived |
| Pitt Freezer Box# |
2 |
| Pitt Freezer Box Grid# |
E5 |
| Available Files |
| EM Picture | Download |
| Final DNAMaster File | Download |
| GenBank File for Phamerator | Download |
