
| Detailed Information for Phage Nonagon |
| Discovery Information |
| Isolation Host | Curtobacterium flaccumfaciens NRRL B-729 SEA |
| Found By | Noelle McDonald |
| Year Found | 2024 |
| Location Found | Pittsburgh, PA USA |
| Finding Institution | University of Pittsburgh |
| Program | Phage Hunters Integrating Research and Education |
| From enriched soil sample? | Yes |
| Isolation Temperature | 30°C |
| GPS Coordinates | 40.43826 N, 79.912968 W Map |
| Discovery Notes | Found next to a sidewalk under leaves and grass clippings.
Plaque picture is after one day incubation.
Annotated by Deborah Jacobs-Sera. |
| Naming Notes | Named after the shape |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Sep 4, 2024 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina NextSeq 1000 |
| Read Type | Single-end reads |
| Read Length | 100 bp |
| Approximate Shotgun Coverage | 6248 |
| Genome length (bp) | 54242 |
| Character of genome ends | Direct Terminal Repeat |
| Direct Terminal Repeat Length | 1289 bp |
| GC Content | 61.2% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | JD |
| Subcluster | -- |
| Cluster Life Cycle | Unknown |
| Other Cluster Members |
|
| Annotating Institution | University of Pittsburgh |
| Annotation Status | In GenBank |
| Plaque Notes | small clear plaques |
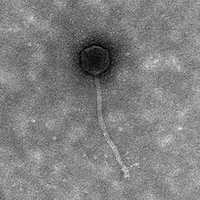
| Morphotype | Siphoviridae |
| Number of Genes | 94 |
| Number of tRNAs | 1 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Minimal DNA Master File | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | PQ868950 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Not in Pitt Archives |
| Available Files |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |