Mycobacterium phage Nonchalant
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Nonchalant | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Gary Liu, Valeria Lopez |
| Year Found | 2025 |
| Location Found | spokane, WA USA |
| Finding Institution | Gonzaga University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 21°C |
| GPS Coordinates | 47.664887 N, 117.39699 W Map |
| Discovery Notes | The phage is discovered from the suface soil.In January , we collected soil samples and discovered a new bacteriophage from it. There was no snow covering the soil. |
| Naming Notes | Phage named "Nonchalant". It wishes the researchers can do the research in calm and without worry. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
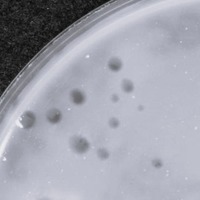
| Lysogeny Notes | No evidence of lysogeny was observed in phage Nonchalant. During host range and stress condition testing, the phage showed: No growth (no plaques) on Gordonia rubi, a non-permissive host. No growth after exposure to harsh environments (pH = 2 and 100 °C), indicating that the phage was inactivated rather than entering a lysogenic state. Clear plaques on M. smegmatis ATCC 607 with an EOP > 1, suggesting strong lytic activity. Nonchalant appears to be a strictly lytic phage under current lab conditions. |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
| Plaque Notes | plaques of Nonchalant have consistent shape and size. The diameter is ≈3mm. |
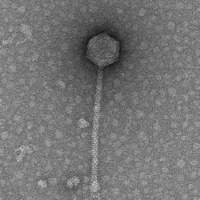
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Not in Pitt Archives |
| SEA Lysate Titer | 1.1x10^10 pfu/mL |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |