Mycobacterium phage Planxty
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Planxty | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Jordan McCarthy |
| Year Found | 2021 |
| Location Found | Gladwyne, PA United States |
| Finding Institution | Saint Joseph's University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 37°C |
| GPS Coordinates | 75.251389 N, 40.045833 W Map |
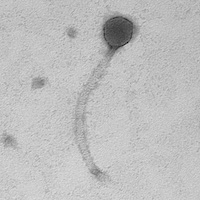
| Discovery Notes | Planxty was collected by Julia Iacovella at Flat Rock Park on 1/25/21. The weather was cold, windy, and sunny with 50% humidity. Julia dug about 15cm into the ground at the edge of the Schuylkill River in between two docks at the park. The soil was under water, so it was very wet and dense. Julia enriched the soil sample, and I performed the subsequent isolation and amplification for the phage. The titer of the High Titer Lysate was 6.2*10^9 pfu/mL. The DNA concentration was 328.5 ng/microliter. Planxty's average capsid size is 50.6 nm and average tail length is 249.7 nm. PCR results using cluster-specific primers suggest that Planxty may be a part of the B3 cluster. |
| Naming Notes | I am an Irish dancer, and my favorite dance is called "Planxty Hugh O'Donnell," so I decided to name the phage "Planxty" as a nickname for that dance. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
| Plaque Notes | After enrichment, Planxty produced total annihilation with direct plating. There was a large plaque on an enriched Spot Test, which was picked for purification plaque streaks. Planxty underwent four rounds of purification and produced clear, circular plaques slightly less than 1mm in diameter with defined edges. These plaques took 24 hours to emerge throughout isolation and amplification. |
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Pitt Freezer Box# | 130 |
| Pitt Freezer Box Grid# | A2 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |