| Detailed Information for Phage Poushou | |
| Discovery Information | |
| Isolation Host | Corynebacterium vitaeruminis CAG1 |
| Duplicates of this Phage | Kayton |
| Found By | Carlos Guerrero |
| Year Found | 2011 |
| Location Found | USA |
| Finding Institution | University of Pittsburgh |
| Program | Phage Hunters Integrating Research and Education |
| From enriched soil sample? | No |
| Isolation Temperature | 30°C |
| GPS Coordinates | 40.44417 N, 79.95306 W Map |
| Discovery Notes | Prophage of Corynebacterium vitaeruminis NCIB 9291. |
| Sequencing Information | |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Nov 15, 2013 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Ion Torrent |
| Approximate Shotgun Coverage | 310 |
| Sanger Finishing Reactions | 14 |
| Genome length (bp) | 40353 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 10 bases |
| Overhang Sequence | CGGCACCCAT |
| End Determination Notes | (ORIGINAL NOTES: No buildups of reads seen from shotgun sequencing.) From the initial Ion Torrent sequencing, there were no obvious coverage dips/increases and/or buildups of read starts/ends, and so this phage's genome was assumed to be circularly permuted. Later Illumina sequencing of other phages in this cluster has shown clearly defined genome ends with a 10-base 3' overhang. The correction to this phage's information and fasta file were made in October 2018. |
| GC Content | 60.0% |
| Fasta file available? | Yes: Download fasta file |
| Characterization | |
| Cluster | EO |
| Subcluster | -- |
| Cluster Life Cycle | Unknown |
| Other Cluster Members |
Click to View |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | In GenBank |
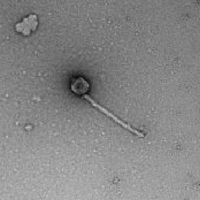
| Morphotype | Siphoviridae |
| Number of Genes | 54 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
Click to ViewPoushou_1 Poushou_2 Poushou_3 Poushou_4 Poushou_5 Poushou_6 Poushou_7 Poushou_8 Poushou_9 Poushou_10 Poushou_11 Poushou_12 Poushou_13 Poushou_14 Poushou_15 Poushou_16 Poushou_17 Poushou_18 Poushou_19 Poushou_20 Poushou_21 Poushou_22 Poushou_23 Poushou_24 Poushou_25 Poushou_26 Poushou_27 Poushou_28 Poushou_29 Poushou_30 Poushou_31 Poushou_32 Poushou_33 Poushou_34 Poushou_35 Poushou_36 Poushou_37 Poushou_38 Poushou_39 Poushou_40 Poushou_41 Poushou_42 Poushou_43 Poushou_44 Poushou_45 Poushou_46 Poushou_47 Poushou_48 Poushou_49 Poushou_50 Poushou_51 Poushou_52 Poushou_53 Poushou_54 |
| Submitted Minimal DNA Master File | Download |
| Publication Info | |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MF197383 |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Available Files | |
| Plaque Picture | Download |
| EM Picture | Download |
| Final DNAMaster File | Download |
| GenBank File for Phamerator | Download |