| Detailed Information for Phage Satis |
| Discovery Information |
| Isolation Host | Streptomyces lividans JI 1326 |
| Former names | Bubbles |
| Found By | Kelly Hartigan |
| Year Found | 2016 |
| Location Found | St. Louis, MO USA |
| Finding Institution | Washington University in St. Louis |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | Not entered |
| GPS Coordinates | 38.644 N, 90.315 W Map |
| Discovery Notes | From dry, mulched soil, 3 cm deep |
| Naming Notes | Satis means "good enough" in latin which has been our mantra throughout this lab. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Feb 21, 2017 |
| Sequencing Facility | The McDonnell Genome Institute at Washington University |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 132 |
| Genome length (bp) | 186702 |
| Character of genome ends | Direct Terminal Repeat |
| Direct Terminal Repeat Length | 1049 bp |
| End Determination Notes | One end (the right) had a clear buildup of reads at a precise base. This was flanking an area of higher-than-average coverage, indicating a terminal repeat, but the other end of the terminal repeat was not well defined. A base was selected for the left end based on approximately where the coverage dropped back to normal, leaving an ~1049 bp terminal repeat. |
| GC Content | 66.7% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | BM |
| Subcluster | -- |
| Cluster Life Cycle | Lytic |
| Other Cluster Members |
|
| Annotating Institution | Washington University in St. Louis |
| Annotation Status | In GenBank |
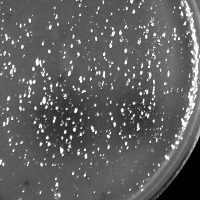
| Plaque Notes | Very small, cloudy plaques with a pinpoint of clearing in the center |
| Number of Genes | 325 |
| Number of tRNAs | 12 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Minimal DNA Master File | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MH576962 |
| Refseq Number | None yet |
| Published in Paper | Shaffer et al 2019, Microbiology Resource Announcements: Complete Genome Sequences of Streptomyces lividans Bacteriophages Satis and JustBecause |
| Archiving Info |
| Archiving status |
Not in Pitt Archives |
| Available Files |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
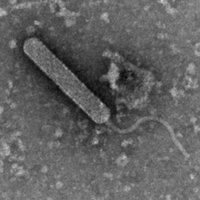
| EM Picture | Download |
| GenBank File for Phamerator | Download |