Mycobacterium phage Zsegetron
Duplicate of Piatt
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Zsegetron | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Allison Darnell |
| Year Found | 2015 |
| Location Found | Holland, MI USA |
| Finding Institution | Hope College |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 32°C |
| GPS Coordinates | 42.787103 N, 86.102207 W Map |
| Discovery Notes | Still in the process of "discovering". From my current data, it is most similar to the K1 phages. |
| Naming Notes | It is named after my high school chemistry teacher Mrs. Szegedi's nickname. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Date Sequencing Completed | May 13, 2020 |
| Sequencing Facility | Hope College |
| Shotgun Sequencing Method | Illumina |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | Unclustered |
| Subcluster | -- |
| Annotating Institution | Hope College |
| Annotation Status | Not sequenced |
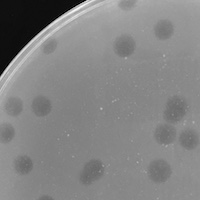
| Plaque Notes | Usually produces plaques around 3-4mm in diameter. Slightly turbid plaques. |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| SEA Lysate Titer | 2.5x10^10 |
| Date of SEA Lysate Titering | Oct 22, 2015 |
| Pitt Freezer Box# | 19 |
| Pitt Freezer Box Grid# | E9 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
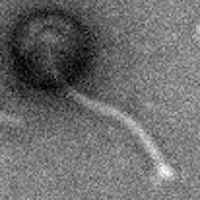
| EM Picture | Download |