| Detailed Information for Phage EniyanLRS |
| Discovery Information |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Duplicates of this Phage | Molarband |
| Found By | Eniyan Kandasamy |
| Year Found | 2015 |
| Location Found | New Delhi, India |
| Finding Institution | Acharya Narendra Dev College |
| Program | Phage Hunters Integrating Research and Education |
| From enriched soil sample? | Yes |
| Isolation Temperature | Not entered |
| GPS Coordinates | 28.539101 N, 77.263726 E Map |
| Discovery Notes | Isolated from a soil sample procured from LRS Hospital, New Delhi in a state called Delhi. The protocol followed for isolation of the Mycobacteriophages was as described in the manual "Phagehunting Teacher Workshop-2012". The work was carried out in Dr.Urmi Bajpai's laboratary at Department of Biomedical Science, Acharya Narendra Dev College, University of Delhi. |
| Naming Notes | The name of the phage is kept by the student Eniyan Kandasamy,Ph.D Student who worked and discovered the Mycobacteriophage from the soil samples as a part of PHAGE DISCOVERY RESEARCH PROJECT (PDRP). The first six letters of "EniyanLRS" are taken from the name of the student:Eniyan and the letters LRS stands for the location in which the soil was obtained. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | May 3, 2016 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 2144 |
| Genome length (bp) | 78536 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 11 bases |
| Overhang Sequence | ACCACTGCAAC |
| GC Content | 56.9% |
| Sequencing Notes | At position 64375 bp 32% of reads contain a G rather than consensus T, at position 75501 bp 33% of reads contain a C rather than consensus T, at position 75509 bp 32% of reads contain a T rather than consensus C. |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | V |
| Subcluster | -- |
| Cluster Life Cycle | Lytic |
| Other Cluster Members |
|
| Annotating Institution | Acharya Narendra Dev College |
| Annotation Status | In GenBank |
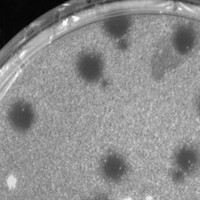
| Plaque Notes | The shape of the plaque which is clear, is circular and the size is about 3-4 mm in diameter. |
| Morphotype | Siphoviridae |
| Number of Genes | 148 |
| Number of tRNAs | 24 |
| Number of tmRNAs | 1 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | KY385381 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Ignore |
| Available Files |
| Plaque Picture | Download |
| Final DNAMaster File | Download |
| GenBank File for Phamerator | Download |
