Mycobacterium phage ILoveMyInterns
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage ILoveMyInterns | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Connor Kunihiro |
| Year Found | 2016 |
| Location Found | Santa Cruz, CA USA |
| Finding Institution | University of California, Santa Cruz |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 37°C |
| GPS Coordinates | 36.983422 N, 122.055944 W Map |
| Discovery Notes | Isolated from a sample taken from a compost pile at the CASFS farm (Center for Agroecology and Sustainable Food Systems) at the University of California, Santa Cruz. The compost contained organic matter, plastic debris, and the occasional rotting mouse carcass. |
| Naming Notes | Named for the gratitude I feel toward my interns that help me with with a project in a research lab at UCSC. Katie and Sammy are wonderful. |
| Sequencing Information | |
| Sequencing Complete? | No |
| Genome length (bp) | Unknown |
| Character of genome ends | Unknown |
| Fasta file available? | No |
| Characterization | |
| Cluster | A |
| Subcluster | -- |
| Cluster Life Cycle | Temperate |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | Not sequenced |
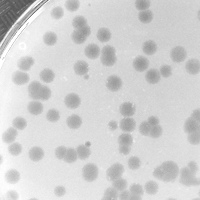
| Plaque Notes | Generally round. Some variation in size. |
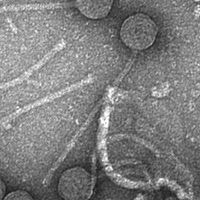
| Morphotype | Siphoviridae |
| Has been Phamerated? | No |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Not in Pitt Archives |
| SEA Lysate Titer | 2600 PFU per microliter |
| Date of SEA Lysate Titering | Nov 3, 2016 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |