| Detailed Information for Phage Lizz | |
| Discovery Information | |
| Isolation Host | Streptomyces viridochromogenes DSM40736 |
| Found By | Jacob Cantrell and Lamia Eldarazi |
| Year Found | 2015 |
| Location Found | St. Louis, MO USA |
| Finding Institution | Washington University in St. Louis |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | 31°C |
| GPS Coordinates | 38.646667 N, 90.314722 W Map |
| Discovery Notes | soil sample collected near bench in Elizabeth Gray Danforth Butterfly Garden. soil: moist depth: 5cm |
| Naming Notes | Lizz was named after Elizabeth Danforth, the namesake of the butterfly garden the soil sample was collected from. |
| Sequencing Information | |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Jun 15, 2020 |
| Sequencing Facility | The McDonnell Genome Institute at Washington University |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 92 |
| Genome length (bp) | 69287 |
| Character of genome ends | Circularly Permuted |
| GC Content | 64.2% |
| Fasta file available? | Yes: Download fasta file |
| Characterization | |
| Cluster | BN |
| Subcluster | -- |
| Cluster Life Cycle | Lytic |
| Other Cluster Members | |
| Annotating Institution | Washington University in St. Louis |
| Annotation Status | In GenBank |
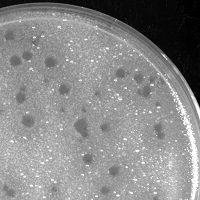
| Plaque Notes | plaques are medium in size and clear. Dark area (halo) surrounding plaques. Some variation in size of plaques on final purification plate. |
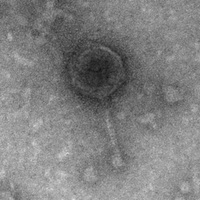
| Morphotype | Siphoviridae |
| Number of Genes | 103 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
Click to ViewLizz_1 Lizz_2 Lizz_3 Lizz_4 Lizz_5 Lizz_6 Lizz_7 Lizz_8 Lizz_9 Lizz_10 Lizz_11 Lizz_12 Lizz_13 Lizz_14 Lizz_15 Lizz_16 Lizz_17 Lizz_18 Lizz_19 Lizz_20 Lizz_21 Lizz_22 Lizz_23 Lizz_24 Lizz_25 Lizz_26 Lizz_27 Lizz_28 Lizz_29 Lizz_30 Lizz_31 Lizz_32 Lizz_33 Lizz_34 Lizz_35 Lizz_36 Lizz_37 Lizz_38 Lizz_39 Lizz_40 Lizz_41 Lizz_42 Lizz_43 Lizz_44 Lizz_45 Lizz_46 Lizz_47 Lizz_48 Lizz_49 Lizz_50 Lizz_51 Lizz_52 Lizz_53 Lizz_54 Lizz_55 Lizz_56 Lizz_57 Lizz_58 Lizz_59 Lizz_60 Lizz_61 Lizz_62 Lizz_63 Lizz_64 Lizz_65 Lizz_66 Lizz_67 Lizz_68 Lizz_69 Lizz_70 Lizz_71 Lizz_72 Lizz_73 Lizz_74 Lizz_75 Lizz_76 Lizz_77 Lizz_78 Lizz_79 Lizz_80 Lizz_81 Lizz_82 Lizz_83 Lizz_84 Lizz_85 Lizz_86 Lizz_87 Lizz_88 Lizz_89 Lizz_90 Lizz_91 Lizz_92 Lizz_93 Lizz_94 Lizz_95 Lizz_96 Lizz_97 Lizz_98 Lizz_99 Lizz_100 Lizz_101 Lizz_102 Lizz_103 |
| Submitted Minimal DNA Master File | Download |
| Publication Info | |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MZ648036 |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Not in Pitt Archives |
| SEA Designator | Lizz |
| SEA Lysate Titer | 2.0x10^10 pfu/mL |
| Date of SEA Lysate Titering | Oct 13, 2015 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |