| Detailed Information for Phage PorkBelly | |
| Discovery Information | |
| Isolation Host | Arthrobacter sulfureus ATCC 19098 |
| Found By | Jose Zamora Lopez, Teams New Genes, Paciphic Rim |
| Year Found | 2025 |
| Location Found | Pasadena, CA USA |
| Finding Institution | University of California, Los Angeles |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 30°C |
| GPS Coordinates | 34.159632 N, 118.125205 W Map |
| Discovery Notes | Taken from outside of a garden. I took it in a shaded area. Depth from which sample was taken (in cm): 3 cm Air temperature if sample collected outdoors (in degrees Celsius): 17.8 C (64 F) Host-Enrichment using Arthrobacter sulfureus (A. sulf), Arthrobacter species (A. sp), Arthrobacter atrocyaneus (A. atro), and Arthrobacter globiformis (A. globi). The soil sample containing the phage was incubated at 30 degrees Celsius for 15 hours with the host bacteria mixture. The mixture was then plated and incubated for 30 degrees Celsius for 48 hours to elicit plaque formation for observation. AL Quarter and Lab section: Instructor: Dr. Tejas Bouklas Teaching Assistant: Pranava Jana Undergraduate Assistant(s): Sarah, Ryan Team #1 Name: Paciphic Rim Team #1 Members: Nolan Reber, Pranay Sashikanth, Hayden Kim, Arman Bhatnagar Team #2 Name: NewGenes Team #2 Members: Marcus Lee, Megan Thanislas, Nick Wilson, Chrissy Tetreault |
| Naming Notes | Marcus’s favorite Korean food |
| Sequencing Information | |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Jan 8, 2026 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina NextSeq 1000 |
| Read Type | Single-end reads |
| Approximate Shotgun Coverage | 344 |
| Genome length (bp) | 49373 |
| Character of genome ends | Circularly Permuted |
| GC Content | 60.0% |
| Fasta file available? | Yes: Download fasta file |
| Characterization | |
| Cluster | FJ |
| Subcluster | -- |
| Cluster Life Cycle | Unknown |
| Other Cluster Members |
Click to View |
| Annotating Institution | University of California, Los Angeles |
| Annotation Status | Being Annotated (Expected completion by 5/1/2026) |
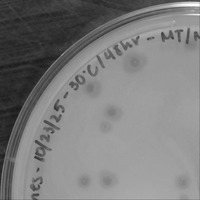
| Plaque Notes | Between plaque assays, morphology varied from pinpoint to clear to halo Description of plaque morphology: Clear, slightly turbid (varies) Average size (and size range) diameter in mm: 2.5-3 (decreased in further weeks, likely decreased titer) |
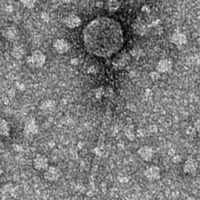
| Morphotype | Siphoviridae |
| Number of Genes | 67 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
Click to ViewPorkBelly_1 PorkBelly_2 PorkBelly_3 PorkBelly_4 PorkBelly_5 PorkBelly_6 PorkBelly_7 PorkBelly_8 PorkBelly_9 PorkBelly_10 PorkBelly_11 PorkBelly_12 PorkBelly_13 PorkBelly_14 PorkBelly_15 PorkBelly_16 PorkBelly_17 PorkBelly_18 PorkBelly_19 PorkBelly_20 PorkBelly_21 PorkBelly_22 PorkBelly_23 PorkBelly_24 PorkBelly_25 PorkBelly_26 PorkBelly_27 PorkBelly_28 PorkBelly_29 PorkBelly_30 PorkBelly_31 PorkBelly_32 PorkBelly_33 PorkBelly_34 PorkBelly_35 PorkBelly_36 PorkBelly_37 PorkBelly_38 PorkBelly_39 PorkBelly_40 PorkBelly_41 PorkBelly_42 PorkBelly_43 PorkBelly_44 PorkBelly_45 PorkBelly_46 PorkBelly_47 PorkBelly_48 PorkBelly_49 PorkBelly_50 PorkBelly_51 PorkBelly_52 PorkBelly_53 PorkBelly_54 PorkBelly_55 PorkBelly_56 PorkBelly_57 PorkBelly_58 PorkBelly_59 PorkBelly_60 PorkBelly_61 PorkBelly_62 PorkBelly_63 PorkBelly_64 PorkBelly_65 PorkBelly_66 PorkBelly_67 |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Not in Pitt Archives |
| Available Files | |
| Plaque Picture | Download |
| EM Picture | Download |