| Detailed Information for Phage Dardanus | |
| Discovery Information | |
| Isolation Host | Gordonia terrae 3612 |
| Found By | Samantha J. Wills |
| Year Found | 2016 |
| Location Found | Fort Myers, FL USA |
| Finding Institution | Florida Gulf Coast University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 30°C |
| GPS Coordinates | 26.460278 N, 81.78 W Map |
| Discovery Notes | My bacteriophage was found in the Food Forest at Florida Gulf Coast University. The soil was dry and sandy. My bacteriophage was both enriched and plated on the bacteria Gordonia terrae. |
| Naming Notes | My bacteriophage is named after the Greek Demigod Dardanus. Dardanus is the son of Zeus and Electra, and the grandson of Atlas according to Greek mythology. This name is meant to reflect my bacteriophage's power. I view my bacteriophage as powerful because it covered my entire plate at the beginning of a serial dilution series. |
| Sequencing Information | |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Jan 16, 2018 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 278 |
| Genome length (bp) | 43143 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 11 bases |
| Overhang Sequence | CCCGGCAGGTA |
| GC Content | 66.6% |
| Sequencing Notes | The sequencing reads from this phage showed that there were at least two deletions in a subset of the population, including one that was present in more than half of the population. The final fasta file that is posted contains these deleted regions, even though the first one is present in a minority of the actual phage particles. Deletion #1 is found in about 70% of the reads, and is a 1218 bp deletion from 25156 to 26373, inclusive. Deletion #2 is found in about 20% of the reads that do NOT have deletion #1, and is 901 bp from 25320 to 26220, inclusive. |
| Fasta file available? | Yes: Download fasta file |
| Characterization | |
| Cluster | CU |
| Subcluster | CU3 |
| Cluster Life Cycle | Temperate |
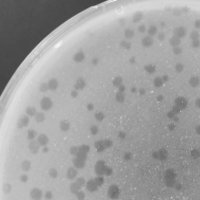
| Lysogeny Notes | My plaques were round and clear. |
| Other Cluster Members | |
| Annotating Institution | University of Louisiana at Monroe |
| Annotation Status | In GenBank |
| Plaque Notes | My phage came from a lyctic plaque inside a turbid plaque. When plated on gordonia terrae for a serial dillution series, small clear plaques developed, as well as large turbid plaques. The least amount of plaques found on a plate was 10 plaques. My plaques are large, however they vary in size. My plaques have a diameter of approximately 2-3.5 millimeters. |
| Number of Genes | 72 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
Click to ViewDardanus_1 Dardanus_2 Dardanus_3 Dardanus_4 Dardanus_5 Dardanus_6 Dardanus_7 Dardanus_8 Dardanus_9 Dardanus_10 Dardanus_11 Dardanus_12 Dardanus_13 Dardanus_14 Dardanus_15 Dardanus_16 Dardanus_17 Dardanus_18 Dardanus_19 Dardanus_20 Dardanus_21 Dardanus_22 Dardanus_23 Dardanus_24 Dardanus_25 Dardanus_26 Dardanus_27 Dardanus_28 Dardanus_29 Dardanus_30 Dardanus_31 Dardanus_32 Dardanus_33 Dardanus_34 Dardanus_35 Dardanus_36 Dardanus_37 Dardanus_38 Dardanus_39 Dardanus_40 Dardanus_41 Dardanus_42 Dardanus_43 Dardanus_44 Dardanus_45 Dardanus_46 Dardanus_47 Dardanus_48 Dardanus_49 Dardanus_50 Dardanus_51 Dardanus_52 Dardanus_53 Dardanus_54 Dardanus_55 Dardanus_56 Dardanus_57 Dardanus_58 Dardanus_59 Dardanus_60 Dardanus_61 Dardanus_62 Dardanus_63 Dardanus_64 Dardanus_65 Dardanus_66 Dardanus_67 Dardanus_68 Dardanus_69 Dardanus_70 Dardanus_71 Dardanus_72 |
| Submitted Annotation in DNA Master Format | Download |
| Submitted Minimal DNA Master File | Download |
| Author List | Download |
| Annotation Cover Sheet | Download |
| Publication Info | |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MN010758 |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Pitt Freezer Box# | 44 |
| Pitt Freezer Box Grid# | G10 |
| Available Files | |
| Plaque Picture | Download |
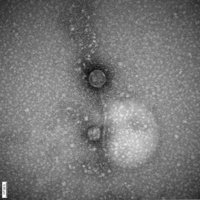
| EM Picture | Download |
| GenBank File for Phamerator | Download |